last update; May. 30. 2022
Help
A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z
| Code | Description |
|---|---|
| IC | inferred by curator |
| IDA | inferred from direct assay |
| IEA | inferred from electronic annotation |
| IEP | inferred from expression pattern |
| IGI | inferred from genetic interation |
| IMP | inferred from mutant phenotype |
| IPI | inferred from physical interaction |
| ISS | inferred from sequence or structural similarity |
| NAS | non-traceable author statement |
| ND | no biological data available |
| TAS | traceable author statemen |
Loose hierarchy of the codes;
[TAS/IDA] > [IMP/IGI/IPI] > [ISS/IEP] > [NAS] > [IEA]
| MR* | Edge |
|---|---|
| MR < 5 | Bold edge |
| MR < 30 | Normal edge |
| MR > 30 | Thin edge |
| Prefix | Type |
|---|---|
| BP | Biological Process |
| CC | Cellular Component |
| MF | Molecular Function |
Evidence code [www.geneontology.org]
| Code | Description |
|---|---|
| IC | Inferred by curator |
| IDA | Inferred from direct assay |
| IEA | Inferred from electronic annotation |
| IEP | Inferred from expression pattern |
| IGI | Inferred from genetic interation |
| IMP | Inferred from mutant phenotype |
| IPI | Inferred from physical interaction |
| ISS | Inferred from sequence or structural similarity |
| NAS | Non-traceable author statement |
| ND | No biological data available |
| TAS | Traceable author statemen |
Loose hierarchy of the codes;
[TAS/IDA] > [IMP/IGI/IPI] > [ISS/IEP] > [NAS] > [IEA]
About logit function in coexpression calculation, please also check our paper.
Obayashi et al. (2018) ATTED-II in 2018: A Plant Coexpression Database Based on Investigation of the Statistical Property of the Mutual Rank Index.
| z score of LS | Edge |
|---|---|
| z > 7 | Bold edge |
| z > 5 | Normal edge |
| z ≤ 5 | Thin edge |
Localization name
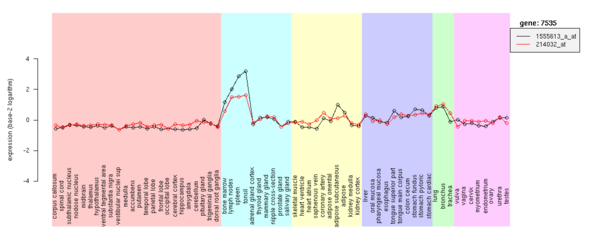
| Species | Accession (Link to NCBI GEO) |
# Of tissues | Averaged # Of reprecation | Reference |
|---|---|---|---|---|
| Human | GSE3526 |
65 | 5.4 | Roth RB et al.(2006) |
| Mouse | GSE1986 |
17 | 1.0 |
After RMA normalization, data were averaged for each tissue. Expression values were base-2 logarithm.
 |
|
| Abbr | name | |
|---|---|---|
| E | E.R. | E.R. |
| G | golg | golgi body |
| K | cysk | cytoskeletal |
| L | lyso | lysosome |
| M | mito | mitochondria |
| N | nucl | nuclear |
| P | plas | plasma membrane |
| S | extr | extracellular |
| V | vacu | vacuole |
| X | pero | peroxisome |
| Y | cyto | cytoplasm |
Localization names joining two abbreviated names with "_", such as "cyto_nucl", indicate dual localization.